Recently I received some emails from users who could not get the correct 3D structure to display in MolView. In this post I want to explain two cases in a bit more detail and show how ambiguous and ill-defined chemical standards cause the 3D structure resolving tool of MolView to be wrong sometimes.
How MolView resolves 3D structures
MolView uses two different webservices to resolve 3D structures. The first step is to convert the structure into an isomeric SMILES representation. SMILES is a text notation that represents chemical structures. These SMILES are generated by a piece of JavaScript that was written as part of Ketcher. When the SMILES is generated, MolView first sends a request to PubChem through their REST API to see if the structure is in their database. If this is the case, MolView will fetch the 3D structure from PubChem. 3D structures from PubChem have been computed using a conformer sampling algorithm (technical references are below). However, if the structure cannot be found in PubChem, MolView will send a request to the Chemical Identifier Resolver to convert the SMILES into a 3D structure. This webservices will use a program called CORINA. This program generates 3D structures by assembling experimental crystal structures.
Case 1: cyclic cis isomers
This case involves cyclic cis isomers. It was brought to my attention that MolView resolves a drawing of cis-1,2-dimethylcyclohexane into trans-1,2-dimethylcyclohexane. Some testing revealed the same issue happens for other simple cyclic compounds. The SMILES that are generated for cis-1,2-dimethylcyclohexane are C1CC[C@](C)[C@](C)C1 when not using explicit hydrogens like in the picture below.
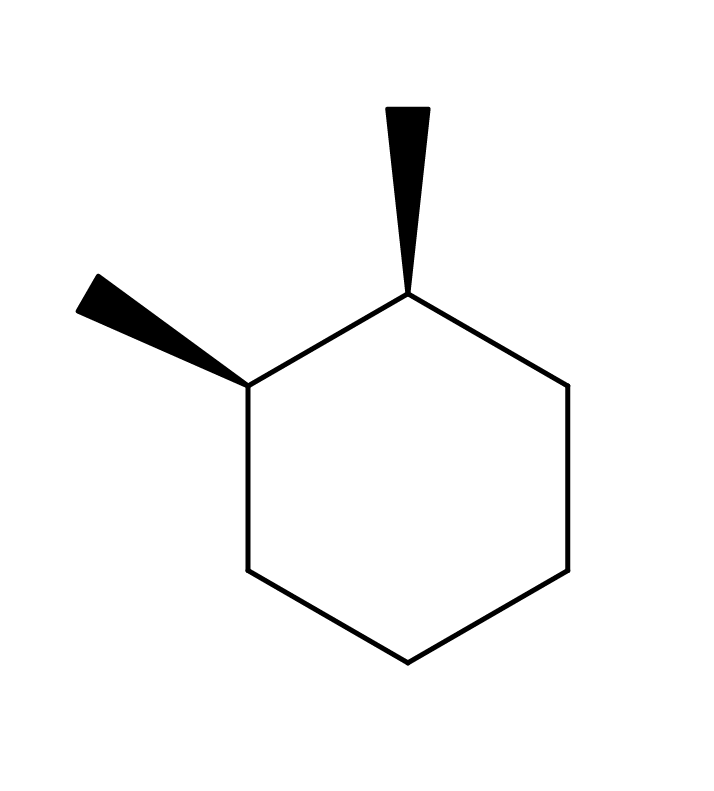
When searching PubChem for this SMILES, we get the entry for 1,2-dimethylcyclohexane, which is the same molecule but without an explicitly defined stereo configuration. The 3D conformation offered by PubChem just so happens to represent the trans isomer (so PubChem has 3 entries for this molecule: cis-1,2-dimethylcyclohexane, trans-1,2-dimethylcyclohexane and 1,2-dimethylcyclohexane). Interestingly enough, the 3D structure is resolved correctly when adding 2 explicit hydrogen atoms as depicted below.
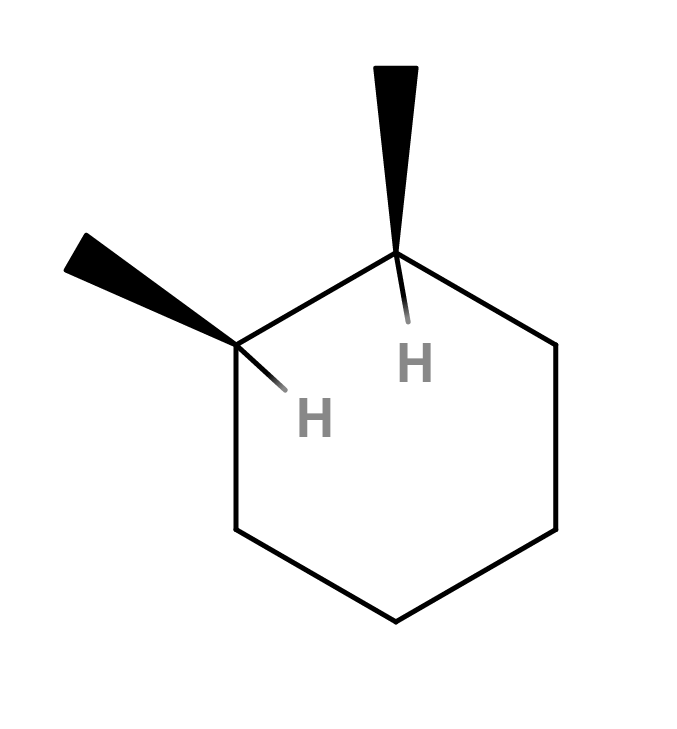
So apparently when those two hydrogen are included in the SMILES (when you add them in the editor they are also added to the generated SMILES), PubChem does return the molecule we are looking for. Therefore, we can say that, at least in this particular case, PubChem interprets the SMILES string generated by MolView in a different way than we would expect. This is a well-known issue of SMILES. Because SMILES is proprietary and not an open project, different chemical software developers have developed different SMILES generation/interpretation algorithms, resulting in different SMILES versions for the same molecule. Therefore, SMILES obtained from different databases or research groups are not always interchangeable unless they used the same software to generate/interpret the SMILES strings. There is now a community effort to create a clear and open specification: http://opensmiles.org/.
Case 2: sulfur tetrafluoride
The second case concerns sulfur tetrafluoride. When you draw SF4 in MolView, the 3D structure provided by PubChem has a tetrahedral geometry (see the 3D conformer on PubChem). This is different from the see-saw structure that is often used for the 3D structure of SF4. This is because the 3D structures from PubChem are generated using a conformer generation/sampling algorithm that is tuned to predict the protein-bound (bioactive) structure of molecules. Therefore, the resulting conformers are often very different from what one would expect for isolated molecules. The difference may become especially noticeable for compounds without direct biological relevance, such as SF4.
If you want to read more about how PubChem works, and how they generate conformers, here are some links:

