I’m writing a draft for MolView 3.0 and I thought it would be awesome to collect some screenshots from old MolView versions. I still didn’t know about git (a program to systematically keep track of your code) when I started building MolView, so some of the old code is unrecoverable. I tried to restore some very old versions from my old laptop and I actually got MolView v0.1 and v0.2 running again for the first time in almost two years! MolView has changed a lot since then and in this post I want to highlight some important developments from the past two years.
Inspiration
You probably used one of these during your chemistry lessons: a molecular model kit. By assembling the plastic parts, you could build all sorts of molecules. We did this in my chemistry class a few years ago to help those who didn’t yet understand how molecular structures looked in three dimensions. It was a lot of fun to finally use our knowledge about molecules to actually build them (unfortunately the parts ran out pretty quickly).
At home I looked for a free, online program to build molecules but I couldn’t find many. However, I did find one particular program: the Virtual Model Kit. This program could be used to draw 2D molecules, convert them to 3D and a lot more which I didn’t really understand back then.
Turning 2D into 3D
I started doing some web development myself around that time and I decided that I could make an even better, more visually appealing program. It took me quite some time to find out how 2D structures could be resolved into 3D structures. The secret turned out to be the Chemical Identifier Resolver which can turn 2D coordinates into 3D coordinates. Unfortunately this service goes down for a couple of days from time to time which is still causing malfunctions in MolView 2.4. For this reason, the Chemical Identifier Resolver will be replaced with a program running on the MolView servers in MolView 3.
Bootstrap
I made MolView available on molview.tk (a domain which was still free back then), showed it to a couple of people and considered the project to be finished. About a half year later I learned about Bootstrap, a user interface framework for websites built by Twitter. It looked quite stylish so I rewrote the user interface of MolView using this toolkit.
More data
Because of the redesign, MolView had gained my attention once again and I implemented a search tool for PubChem (small molecules) and RCSB (proteins). In the mean time the design got more complicated and Bootstrap was not really suitable anymore so I developed a new, handwritten design to seamlessly fit all features in one interface (for some reason black became the primary color).
A Top-Level Domain
The 1st of July 2014 is the day that molview.org was launched. This new domain contained the first version of the 2.x series. One of the most exciting new features in this version is the integration of the Crystallography Open Database enabling users to view crystal structures. I also discovered git and version control (keeping track of your code) around this time. MolView 2.1 (basically the same as v2.0) is the first version which is safely stored on GitHub.
An autocomplete
I improved MolView a lot during the summer of 2014. MolView got a much whiter design and an autocomplete feature to quickly find proteins, small molecules and crystal structures. This is particularly useful for the crystal structures because the Crystallography Open Database does not have a search index (therefore searching it is relatively slow). The protein collection which you can find via the autocomplete is derived from the Molecular Machinery Poster from the Protein DataBank.
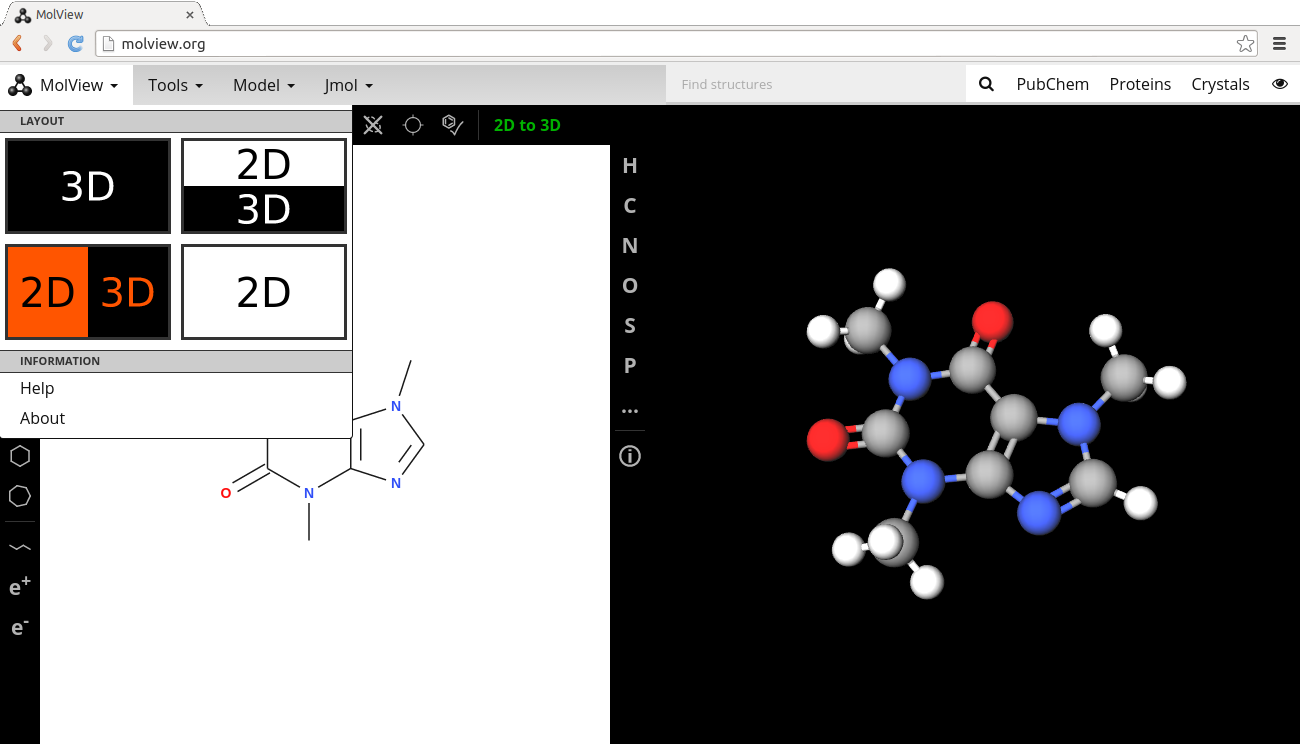
A new sketcher
The sketcher of MolView has changed twice. I first switched from a program called Ketcher to a program called MolEdit because Ketcher caused conflicts with some other parts of MolView. MolEdit is developed by Molsoft for academic and personal use. I contacted Molsoft and initially they agreed that I was allowed to use MolEdit as long as there was a link to their website. But just a few weeks before the planned release of MolView 2.2 (12 November) I was contacted by Molsoft and asked to put a Molsoft brand right on top of the sketcher. I didn’t quite agree with that (I don’t want advertisement of any form in MolView!) and decided to try the impossible; writing a molecule sketcher from scratch in only a couple of weeks. Today MolView has, in my opinion, one of the best molecule editors on the internet!
A new logo
Recently a new logo for MolView was announced. You have probably read about it in the previous post.


